I'm dressed in a white coat and nitrile gloves and there are no optics in sight. To most, this might seem like some of the more unusual birding attire, and yet I find myself within inches of something special – Siberian Stonechat. Admittedly, not the bird itself but its droppings.
As a PhD student working under Prof Martin Collinson in the wildlife forensics lab at the University of Aberdeen, this is the ninth time I have found myself in this scenario in autumn 2019, with an 'eastern' stonechat in my hands just begging to be identified. Admittedly, after the poo hit the fan with the Sennen Paddyfield Pipit identification in December, I welcome this return to something a little more familiar.
I feel very lucky to be tasked with conducting genetic identifications of tricky vagrants from Britain and across the globe. Our 'greatest hits' – birds whose identification has been based on or confirmed by DNA – include British firsts of Pale-legged Leaf Warbler, Eastern Yellow Wagtail, 'Eastern' Grasshopper Warbler and Siberian and Stejneger's Stonechats, as well as Acadian Flycatcher, Western Subalpine, Eastern Subalpine and Moltoni's Warblers – and quite a few firsts for other European countries, too.

Cheshire’s first eastern stonechat appeared to show a mix of characters in the field depending on light and other factors; DNA analysis confirmed it as Siberian Stonechat (Mike Atkinson).
In the genes
Almost all birders will be aware of genetic identifications creeping increasingly onto the rarity radar in the last 10 years or so. If we are lucky, then a rare bird will have the decency to find its way into a mistnet, and any feathers that happen to be dislodged during normal handling as the bird is ringed can be collected and sent for genetic testing.
In most cases, though, vagrants are generally located in the field and not the net. However, we can get DNA from just about any biological material. While attempting to reach an identification, a watchful eye may see a feather or two dislodged as a bird preens, a pellet regurgitated or, as on one memorable occasion last autumn, the bird may bleed onto an accessible twig. More often than not, though, the easiest means of collecting a sample is to simply wait for the bird to defecate, spot where the faeces lands and move in to collect when the bird vacates the area.
That's when the fun starts. Faecal matter is probably the poorest medium from which we can recover DNA in the lab. The first issue that arises is whether the right dropping has been collected. The likelihood is that the bird you are watching won't have been the first to have answered nature's call from that particular perch, and there is nothing more frustrating than watching a subalpine warbler ID slip away as the DNA comes back as a match for Dunnock.
Once confident that the correct deposit has been identified, how best to collect it? We have found samples that have been collected and allowed to dry naturally work best. As such, collection in an absorbent material, such as a tissue, a scrap of paper from a notebook or even a sheet of cigarette paper, is ideal. This keeps the sample intact as it dries. Ultimately, you have to make the most of what you have to hand, but direct collection into a zip-lock bag or similar can lead to the sample breaking up into a fine statically charged powder once dry, which is both difficult to work with and potentially a contamination risk.

The lengths some birders go to! Poo samples were collected to confirm the Cheshire eastern stonechat as Siberian (@MarburyPatch).
The DNA within a faecal sample comes from cells dislodged from the digestive tract of the bird. Unfortunately, digestive enzymes are excellent at breaking down DNA. The result of this is that the DNA we can recover will tend to be broken into tiny pieces which are too short for our standard gene amplification techniques, and in some cases can be so fragmented as to be unusable. As such, extra time is required to design, test and optimise bespoke protocols for target taxa.
It is because of all these constraints that we say there is only a 50% chance of successfully generating an ID from a faecal sample. This would compare with 99% confidence from feather, blood and tissue samples.
Labwork
On receiving a sample in the lab – I am one of very few people for whom receiving poo in the post constitutes a good day – we start the process of isolating the DNA from the sample. This is done by placing the dropping overnight into a digest of an enzyme called Proteinase K. This is the molecular equivalent of leaving your dishes to soak. It breaks down proteins and organic materials, releasing DNA from within the cells. From here it is a short process to filter out the waste material, leaving behind pure DNA. For faecal samples, this means we are left with the DNA of the bird and whatever it has been eating. In most cases this isn't an issue, but if the sample is from a bird that eats other birds, for example Northern Harrier, it can be challenging to specifically target the DNA of either predator or prey.
There are several ways of taking the tiny amounts of DNA we may retrieve from a sample and turning it into an identification, but 'on the cheap', which is how we have to do things, the easiest way is to replicate and isolate individual genes for sequencing. Our standard procedure with higher-quality blood and feather samples is to attempt to sequence whole genes using very generic protocols that work with almost all bird species.
However, the highly fragmented nature of faecal samples mean that we are forced to target very small pieces of the genome with known differences between the taxa of interest and must tailor our protocols to be specific to both the particular fragments and relevant taxa. This is why it takes us longer to process faecal samples than feathers.

Not your usual birding! The team at the University of Aberdeen – left to right: Thom Shannon, Becky Shaw, Martin Collinson and Tess Senfeld – get down to work (top), testing the DNA from feather samples (Thom Shannon).
In the case of stonechats we sequence a short stretch of the mitochondrial 'ND2' gene. But very short stretches are often enough. For a sense of scale, birds have around 1 billion base pairs of DNA in every cell, and yet all that is required to identify whether we are dealing with a Stejneger's, Siberian or European Stonechat is a 250 base stretch of DNA from a single gene.
Once the sequence has been acquired, a search of a publicly accessible online DNA sequence database is all that is required. This compares your sequence with the entire database and will return the specimens that provide the closest matches along with a percentage similarity to your sequence. With a little critical analysis, taking into consideration how similar your sequence is to the closest matches and whether all relevant taxa are present in the database, we can settle on an identification.
The process from poo to ID is a fairly long one – in the order of days rather than hours. However, it is worth the effort of both the labbies and the eagle-eyed poo collectors in the field. In a number of cases identification would remain indefinitely unresolved without the addition of genetic information.
Eastern stonechats
One species pair that is reliant on genetic analyses are the 'eastern stonechats'. The BBRC currently doesn't accept these to species level without a genetic identification. Siberian Stonechats are usually found from the eastern edge of European Russia east to Lake Baikal and south to Afghanistan and north-west Mongolia. The range of Stejneger's Stonechat is further east, encompassing eastern Siberia, Japan, Korea, north-eastern China and eastern Mongolia.
Given the ranges of these two species, it would seem logical that Siberian Stonechat would be the more common visitor to Britain, but is that a safe assumption? In the wake of a bumper autumn, we were sent samples from nine stonechats with the task of conclusively identifying each to species. Of these seven were faecal samples and two were from feathers.

Shetland’s first and second Stejneger's Stonechats were recorded in autumn 2019, with this bird on Mainland in October (Penny Clarke).
The first sample came from Brake, Shetland. Present from 28 September until 6 October, this bird was suggested as a Siberian in the field. It was quickly followed by a female putative Siberian at Spurn on 6 October. Fortunately, this bird was trapped and ringed that evening, providing a single dropped feather. It was seen briefly the following morning before disappearing in worsening weather. Both birds shared the classic pale and frosty appearance that is typical of Siberian Stonechats in autumn, so it was perhaps unsurprising that both were confirmed as maurus genetically.
The next three samples were all found to be carrying Stejneger's DNA. All of these birds were darker and more deeply saturated in appearance, particularly on the upperparts. Two of them came from Shetland. Birds caught and ringed at Westing on Unst (8-14 October) and Leebitten on Mainland (17-28 October) became the first and second records of Stejneger's for the islands. Whitburn, Co Durham, provided the venue for the third sample between 22 and 24 October.
Spurn's second eastern stonechat of the autumn proved to be the most divisive. Frequenting Peter's Lane between 9 and 13 November, this bird was on the paler end of the spectrum with a notable unmarked two-toned rump. These features led some observers to suggest it was another Siberian. However, the bird lacked the classic 'cold' tone produced by pale buff fringes to the mantle feathers, giving it a somewhat warmer appearance more in keeping with Stejneger's. Described as “a real chameleon of a bird” by Spurn Bird Obs, the bird seemed to give wildly varying impressions dependent on viewing conditions. In the end the DNA proved to be the conclusive feature, as this became the lab's fourth Stejneger's identification of the season.

The Whitburn, Co Durham, bird – present from 22-24 October 2019 – was confirmed to be carrying Stejneger's DNA (Martyn Sidwell).
Wintering birds
The final two samples collected were from long-staying wintering birds. The first was a first-winter male at Hollesley Marshes in Suffolk (present 1 Dec-11 Jan). This individual was pale and frosty with a large, lozenge-shaped pale rump. The bird was called as maurus from the outset and this was again backed up by its DNA.

This stonechat at Hollesley Marshes RSPB, Suffolk, lingered into early 2020 and was confirmed as Siberian by DNA analysis (Andrew Moon).
On the opposite side of the country, Cheshire got an early Christmas present in the form of its first eastern stonechat, at Ashton's Flash from 24 December until 11 March (at least). Not unlike the Peter's Lane bird, this individual seemed to show mixed features in different conditions. Benefiting from its prolonged stay, a better consensus was built up from observers in the field and this lent towards maurus. This was reflected in the DNA result.
We ended up with four of each species, suggesting that the more easterly Stejneger's may not be as rare a vagrant to Britain as previously thought. A ninth sample, a bird found on Fair Isle, was a damning reminder that faecal samples are not guaranteed to be a success. After unfortunately failing to yield any usable DNA from two different droppings, this was the one that got away.
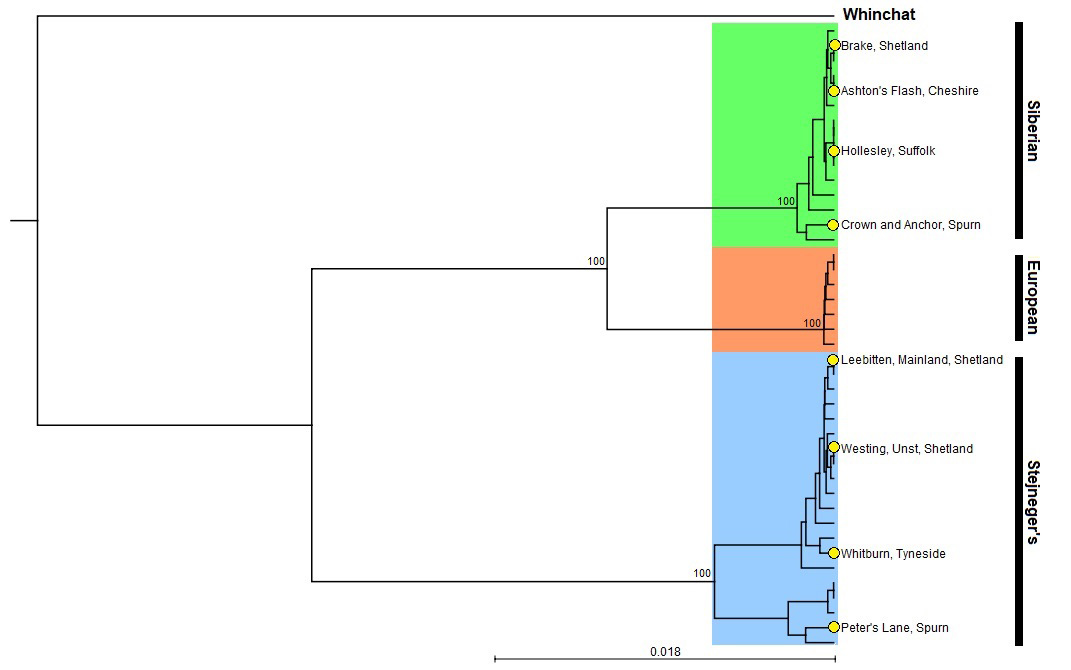
A phylogenetic tree showing the relative position of the eight 'eastern' stonechat samples genetically identified to species during autumn 2019.
Not all birders are enamoured of genetic identifications becoming routine, feeling that birding is becoming too technical. In fact, there is some hope that this burst of genetics may eventually confirm that the majority of Siberian and Stejneger's Stonechats occurring here can be identified in the field. From these birds, and eastern stonechats we have identified in previous years, it has become apparent that for the most part, the field identifications tended to be correct.
The occurrence of ambiguous forms such as the bird at Peter's Lane means that the need for genetic analysis of some birds persists, but we may find that for many, perhaps even most, individuals, it should be possible to propose a firm identification without genetic input. With an increase in genetically confirmed individuals, it is hoped that diagnostic field characters might be gleaned to separate these difficult intermediate birds.
So, it turns out one bird's waste, is another birder's treasure. From a single dropping we are able to make an identification on an ambiguous bird, and from there attempt to facilitate birders in the field in the future. Our work is far from an alternative to traditional fieldcraft, but a potentially powerful supplement to it.
- This article was originally published in the April 2020 issue of Birdwatch magazine.




